
Cell death is an essential part of development and normal adult homeostasis. Failure to maintain this homeostasis can have disastrous consequences. Too much apoptosis can lead to any number of degenerative diseases. Too little apoptosis will result in cancer. We crossed two models of apoptosis into the DGRP and identified nearly 100 candidate modifiers of apoptosis (Palu et.al 2019, BioRxiv). We are characterizing these modifiers through an established pipeline to try and determine how these genes are altering apoptosis.
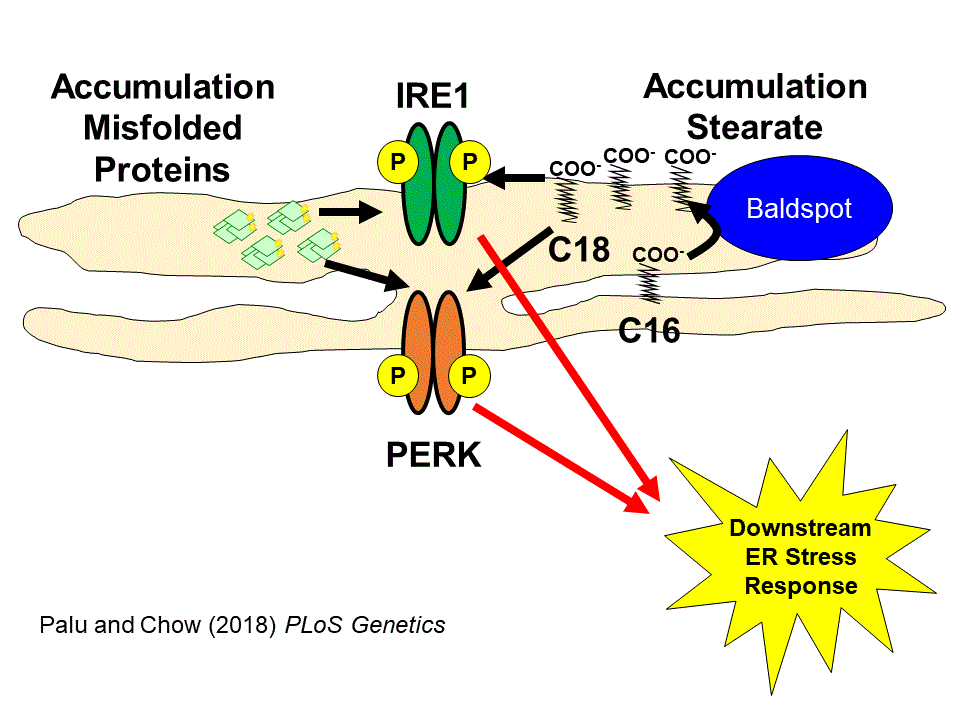
The Drosophila fatty acid elongase Baldspot alters the balance of palmitate and stearate in the cell to modify the response to endoplasmic reticulum stress (Palu and Chow 2018, PLoS Genetics). Given the previous associations between fatty acid accumulation, ER stress, and metabolic disease, we want to explore the role Baldspot plays in regulating metabolic homeostasis after dietary manipulation or ER stress. Preliminary data suggests that loss of Baldspot in the adipose/liver tissues sensitizes the fly to a high sugar diet while rendering them resistant to a high palmitate diet. We plan to study the role of Baldspot in maintaining physiological homeostasis as well as the role ER stress plays in challenging this process.
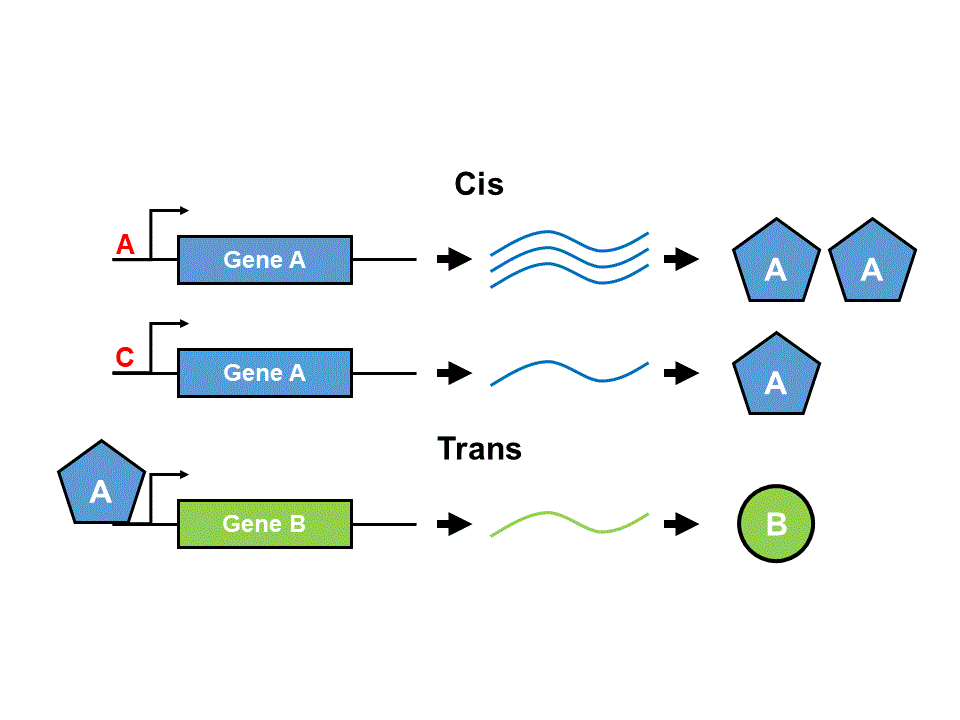
Earlier work used the DGRP to identify modifiers of ER stress using genome-wide association analyses (Chow et.al 2016, HMG). These analyses, however, can only identify genetic modifiers that are close in proximity (“cis”) to the causative SNP. We would not identify any modifiers that are far from (“trans”) that SNP. We plan to use transcriptional variation to identify modifiers that function in trans to the genetic variation present in the DGRP. As in the previous study, we overexpressed misfolding protein in the precursor tissue to the adult Drosophila eye: the eye imaginal disc. We then isolated this tissue from 130 DGRP lines for which we also have phenotypic data and performed RNA-seq. We hope to mine this data for new modifiers of ER stress.
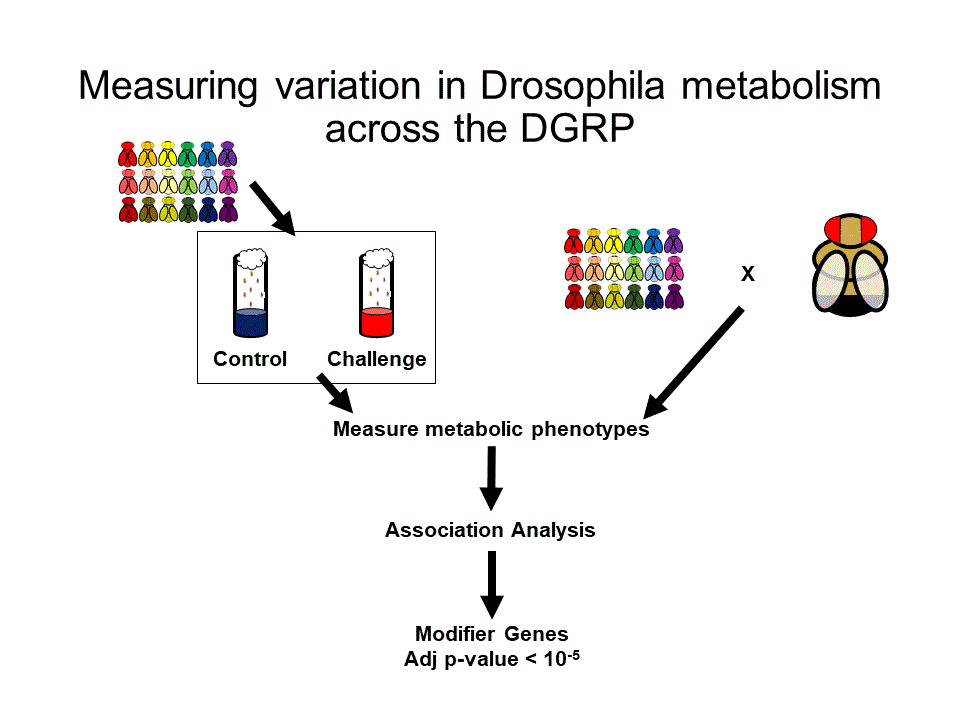
This is the newest project in the Palu lab, and involves taking advantage of the natural genetic variation present in the DGRP to identify candidate modifiers in models of metabolic disease. We will explore a number of dietary (i.e. high sugar, high fat) as well as genetic (sir2-/- or AKHR-/-) models of metabolic dysfunction. We hope to initially answer two questions. How does the model of metabolic disease impact the modifiers we identify? What about the type of measurement taken (glucose or triglycerides)? We then hope to characterize the functions of these modifiers in a variety of tissues and physiological contexts.



